Welcome to nlfem’s documentation!
Documentation
The implementation has a Python interface and a C++ core. The Python interface provides the main functionality. The C++ core documentation should provide sufficient insight to enable the user to extend nlfem, for example, by new kernel functions.
Build and Install
Docker
To quickly set up a terminal with an nlfem installation execute
docker run --interactive --tty registry.gitlab.uni-trier.de/pde-opt/nonlocal-models/nlfem:latest bash
The above image contains a Jupyter notebook installation. You can start a notebook which can be reached
at http://localhost:<port> by
docker run -p <port>:80 registry.gitlab.uni-trier.de/pde-opt/nonlocal-models/nlfem:latest jupyter notebook
Manual Installation
Ubuntu 22.04 LTS
On Ubuntu the required packages can be installed by the following commands:
sudo apt-get update
sudo apt-get install git gcc g++ libarmadillo-dev liblapack-dev libmetis-dev python3-venv python3-dev libgmp-dev libcgal-dev
mkdir nlfemvenv
python3 -m venv nlfemvenv/
source nlfemvenv/bin/activate
The terminal will now indicate that you are in (nlfemvenv). In order to install nlfem run:
git clone https://gitlab.uni-trier.de/pde-opt/nonlocal-models/nlfem.git
cd nlfem
python3 -m pip install -r requirements.txt
python3 setup.py build install
Required packages
In general a system has to meet the following requirements.
The basic requirements are the programs
gcc, g++, python3-dev, python3-venv, libgmp-dev, libcgal-dev, metis,libmetis-dev, libarmadillo-dev. You further need the Python 3 packagesnumpyandscipy. If you want to change the Cython code, you require the packageCython.For running the examples you additionally need to install the Python packages
matplotlib, meshzoo. Make sure to install the correct versions by using therequirements.txt.
License
nlfem is published under GNU General Public License version 3. Copyright (c) 2021 Manuel Klar, Christian Vollmann
Quick Start
To test the rates for the constant kernel run
cd nlfem/examples/test2D
python3 computeRates2D.py -s 4 -f testConfConstant
Run a test with other configurations by indicating the file e.g. -f testConfFull. The option -s indicates the number of refinement steps.
Usage
Let \(\Omega \subset \mathbb{R}^{d}, (d=1,2,3)\) be a bounded domain and \(\Gamma^D \subset \Omega^c\), where we allow that \(\Gamma^D = \emptyset\). The package nlfem allows to assemble stiffness matrices which discretize the nonlocal operator
where \(\mathbf{C} \in \mathbb{R}^{n\times n}, (n \geq 1)\) is a possibly matrix-valued integral kernel and \(B_{\delta}(\mathbf{x})\) a \(p\)-norm ball (\(p \in [1, \infty]\)) of radius \(\delta\). Details about the mathematical background of nlfem are presented in [Klar2022].
Example We exemplify the usage of nlfem by solving a scalar, nonlocal Dirichlet-type problem
where \(\Omega\) by a 2d-disk of radius 0.9 with a nonlocal boundary \(\Gamma^D\) of width \(\delta=0.1\).
Finite element mesh
The construction requires a finite element mesh in a 1d, 2d, or 3d domain. The mesh is characterized by its elements and vertices. The elements are given by a numpy.ndarray of datatype np.int_ and shape (nE, d+1), where nE is the number of elements and d the dimension of the domain. The vertices are a numpy array of floats with shape (nV, d), where nV is the number of vertices.
The domain described by the the above arrays needs to be divided into different parts according to their purpose. To that end we define a numpy nd.array called elementLabels of type np.int_ and length nE. It assigns a label to each element. Negative elements indicate the Dirichlet boundary, any positive element is considered a part of the domain and zero labeled elements are not part of the integration.
Example The finite element mesh
(that is vertices and elements) of the
required form can be obtained for example from gmsh.
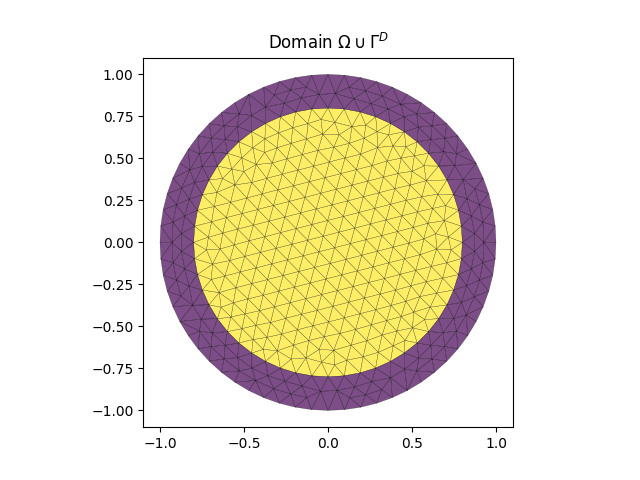
In the example the elementLabels are set to -1 on \(\Gamma^D\) (blue in the below figure) and 1 on \(\Omega\) (yellow).
The plot is based on a domain saved in examples/docs/mesh.pkl.
import pickle
import matplotlib.pyplot as plt
mesh_arrays = pickle.load(open("mesh.pkl", "rb"))
elements, vertices, elementLabels = mesh_arrays["elements"], mesh_arrays["vertices"], mesh_arrays["elementLabels"]
plt.tripcolor(vertices[:, 0], vertices[:, 1], elements, facecolors=elementLabels, edgecolors="black", linewidth=.2, alpha=.7)
plt.title(r"Domain $\Omega \cup \Gamma^D$")
plt.gca().set_aspect('equal')
plt.savefig("domain.png")
plt.close()
Settings
The kernel is a dictionary which contains the keys function, horizon, outputdim and possibly fractional_s.
A list of implemented kernel functions can be found via show_options().
The horizon determines the interaction radius of the kernel and outputdim tells whether the kernel is scalar or matrix-valued. Given the kernel has a singularity which requires regularizing integral transformations we need to determine the parameter s of the singularity in fractional_s. The quantity is required for the special integration routines fractional and weakFractional which need to be applied to those kernels.
Example We choose a scalar kernel of truncated fractional type
where \(d=2\) and \(s=0.4\). The corresponding dictionary reads as
kernel = {
"function": "fractional",
"horizon": 0.1,
"outputdim": 1,
"fractional_s": 0.4
}
As optional parameter it is possible to hand over varying kernel coefficients with the key "Theta". Of course, the implemented kernel has to support the usage of the given information.
This is so, for example, for the kernel functions "theta" and "sparsetheta" which
evaluate
and
respectively. The coefficients Theta are expected to be of type scipy.sparse.csr_matrix. However, there are no other restrictions, so that any use of the coefficients inside of the kernel function is possible.
Note also that new kernels can be defined.
The forcing-term has a similar structure and contains a key function indicating the forcing term.
Again, a list of implemented kernel functions can be found via show_options().
Note that the assembly of the right hand side is standard and nlfem offers this functionality for convenience only.
Example We choose \(f(x) = -2(x_1+1)\) which can be selected by
forcing = {"function": "linear"}
All other settings are stored in the dictionary conf.
The truncation routines are selected by the method given in approxBalls.
The quadrature rule which is used in the truncation routine is indicated by numpy arrays of quadrature points and weights.
For kernels which exhibit a singularity it is possible to select another integration routine specifically for touching elements
(e.g. fractional, weakFractional). However, the choice does not affect the quadrature for nonsingular kernels.
The function show_options() prints a list of the singular kernels.
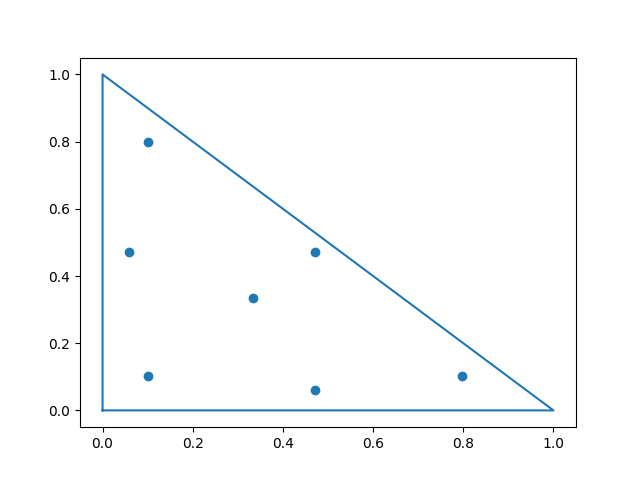
Example We discretize the problem with a continuous Galerkin ansatz space, and use a retriangulation with caps to approximate
the truncation of the interaction set. We choose a seven-point quadrature rule with points Px and weights dx which is advantageous for the approximation of the truncated interaction set of a finite element, as the points lie close to the edges of the element [DElia2021].
import numpy as np
Px = np.array([[0.33333333333333, 0.33333333333333],
[0.47014206410511, 0.47014206410511],
[0.47014206410511, 0.05971587178977],
[0.05971587178977, 0.47014206410511],
[0.10128650732346, 0.10128650732346],
[0.10128650732346, 0.79742698535309],
[0.79742698535309, 0.10128650732346]])
dx = 0.5 * np.array([0.22500000000000,
0.13239415278851,
0.13239415278851,
0.13239415278851,
0.12593918054483,
0.12593918054483,
0.12593918054483])
The example configuration is set by the dictionary:
conf = {
"ansatz": "CG",
"approxBalls": {
"method": "retriangulate"
},
"quadrature": {
"outer": {
"points": Px,
"weights": dx
},
"inner": {
"points": Px,
"weights": dx
},
"touchingElements": {
"method": "fractional",
"ntensorGaussPoints": 4
}
},
"verbose": True
}
Note All options for the settings in kernel, function and conf can be printed by nlfem.show_options().
Empty dictionaries in the required form can be obtained from nlfem.get_empty_settings().
Assembly
Given the above settings the assembly of the nonlocal stiffness matrix is effected via
import nlfem
mesh, A = nlfem.stiffnessMatrix_fromArray(elements, elementLabels, vertices, kernel, conf)
f = nlfem.loadVector(mesh, forcing, conf)
The dictionary mesh contains information about the labeling of elements, vertices and dof.
The labels of the degrees of freedom depend on the kernel (scalar or matrix-valued) and the ansatz space (continuous (CG) or discontinuous (DG) P1 elements).
Therefore the function nlfem.stiffnessMatrix_fromArray() automatically deduces vertexLabels and dofLabels.
A vertex is labeled according to the labels of the elements it belongs to and it is given the smallest of those element labels.
The dof labels are identical to the vertexlabels for CG ansatz spaces and scalar-valued kernels.
For DG ansatz spaces the labels are directly obtained from the element labels.
The labels of the degrees of freedom on the domain \(\Omega\) and the Dirchlet domain \(\Gamma^D\) are derived from the element labels and stored in mesh as dofLabels.
Omega = mesh["dofLabels"] > 0
Gamma = mesh["dofLabels"] < 0
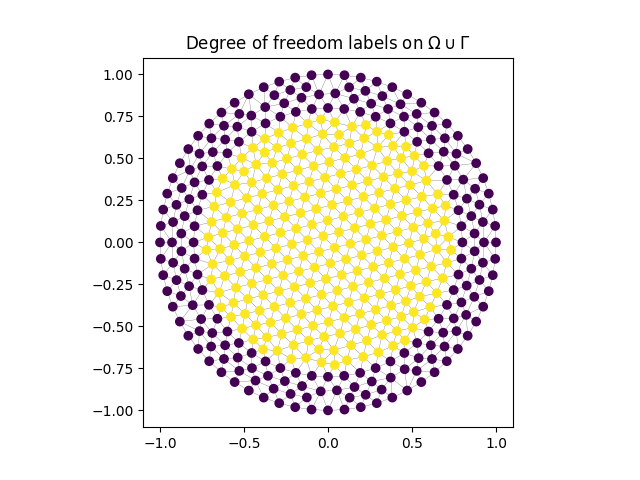
Example In the given example we have a CG ansatz space and a scalar-valued kernel so that the dofLabels are identical to the vertexLabels.
plt.triplot(vertices[:, 0], vertices[:, 1], elements, color="black", linewidth=.2, alpha=.7)
plt.scatter(vertices[:, 0], vertices[:, 1], c=mesh["dofLabels"])
plt.title("DoF labels")
plt.gca().set_aspect('equal')
plt.savefig("doflabels.png")
plt.close()
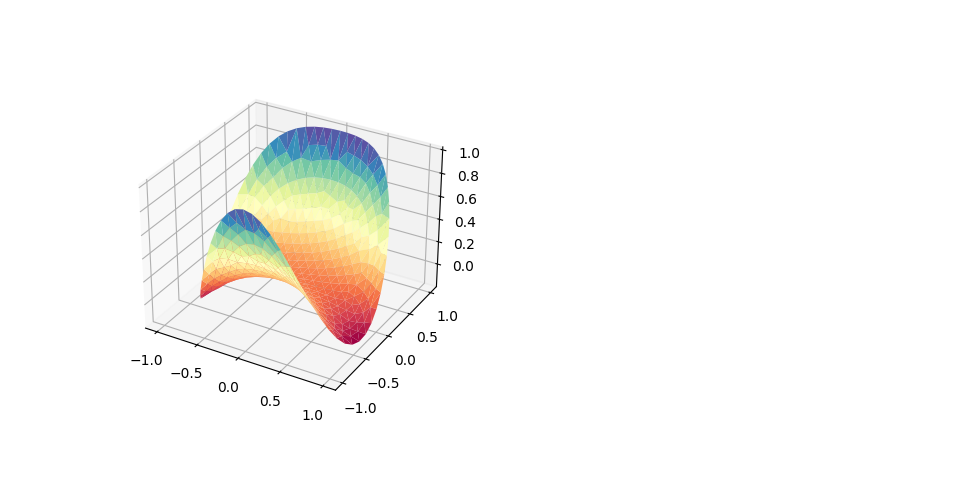
The solution of a Dirichlet-type problem requires the definition of Dirichlet data. The function below implements \(g(x) = x_1^2 x_2 + x_2^2\).
def g(vertices):
return vertices[:, 0] ** 2 * vertices[:, 1] + vertices[:, 1] ** 2
Ultimately we can solve the nonlocal Dirichlet-type problem via
from scipy.sparse.linalg import cg
f_O = f[Omega]
A_OO = A[Omega][:, Omega]
A_OD = A[Omega][:, Gamma]
g_D = g(vertices[Gamma])
f_O -= A_OD.dot(g_D)
u = np.zeros(vertices.shape[0], dtype=float)
u[Omega] = cg(A_OO, f_O)[0]
u[Gamma] = g_D
fig = plt.figure(figsize=plt.figaspect(0.5))
ax = fig.add_subplot(1, 2, 1, projection='3d')
surf = ax.plot_trisurf(vertices[:, 0],vertices[:, 1], u, cmap=plt.cm.Spectral)
plt.savefig("solution.png")
plt.close()
Defining integral kernels
New kernels can be added to nlfem in the following way. Declare your kernel kernel_myKernel() in src/model.h and implement it in src/model.cpp. You kernel function has to exactly meet the interface of the
function pointer model_kernel.
void (*model_kernel) (const double * x, long labelx,
const double * y,long labely,
const MeshType &mesh, double * kernel_val);
In order to be able to choose the kernel in the Python interface add an entry
{"myKernelName", kernel_myKernel}
to the map lookup_kernel in src/Cassemble.cpp.
D’Elia, M., Gunzburger, M., & Vollmann, C. (2021). A cookbook for approximating Euclidean balls and for quadrature rules in finite element methods for nonlocal problems. Mathematical Models and Methods in Applied Sciences, 31(08), 1505-1567.
Klar, M., Vollmann, C., & Schulz, V. (2022). nlfem: A flexible 2d Fem Code for Nonlocal Convection-Diffusion and Mechanics. arXiv preprint arXiv:2207.03921.
Python interface
- nlfem.show_options()
Shows options for kernel, load and configuration settings. The kernel and forcing functions are imported directly from src/Cassemble.cpp.
- nlfem.get_empty_settings()
Returns empty dictionaries which can be filled with values. Possible options can be printed via show_options().
- nlfem.stiffnessMatrix_fromArray()
Computes the stiffness matrix corresponding to the nonlocal operator
\[-\mathcal{L}(\mathbf{u})(\mathbf{x}) = 2 \int_{B_{\delta}(\mathbf{x}) \cap \widehat{\Omega}}(\mathbf{C}_\delta(\mathbf{x}, \mathbf{y}) \mathbf{u}(\mathbf{x}) - \mathbf{C}_\delta(\mathbf{y}, \mathbf{x})\mathbf{u}(\mathbf{y})) d\mathbf{y},\]on a given mesh. This function is a wrapper around
stiffnessMatrix().- Parameters:
elements – Numpy array containing the information about the elements. The values in the array
"elements"are expected to be of datatypenumpy.int.elementLabels – The values in the array
"elementLabels"are expected to be of datatypenumpy.int. Elements in the domain have positive labels. Elements in the nonlocal Dirichlet boundary have negative labels. For this purpose it does not matter which positive or negative number is used, and the kernels can depend on the element labels. The routine does not compute contributions of elements with label zero, i.e. they are ignored. The label zero can therefore be added to connect disconnected domains. The labels of the vertices are automatically derived from the element labels. In the case of Discontinuous Galerkin this means that the signs of the element labels and corresponding vertex labels coincide. In case of Continuous Galerkin this means that a vertex gets the smallest label of the non-zero labels of the adjacent elements.vertices – The values in the array
"vertices"are supposed to be of typenumpy.float64.kernel – The kernel is assumed to be a dictionary containing the keys
"function", and"outputdim". The value of"function"is a string which contains the name of the kernel (e.g. “constant”). You find all available options in the function src/Cassemble.cpp/lookup_configuration(). The key"outputdim"describes the output dimension of the kernel function. In case of a scalar valued kernel this would be 1. In case of the kernel “linearPrototypeMicroelasticField” this would be 2 (find an example in examples/Test2D/testConfFull.py). Note, that the value of"outputdim"does not define the output dimension of the kernel, but describes it. The value has to be consistent with the definition of the corresponding kernel in src/model.cpp.configuration – Dictionary containing the configuration (find an example in examples/test2D/conf/testConfFull.py).
- Return mesh:
Dictionary containing the mesh information. Can also be used for
stiffnessMatrix()andloadVector().- Return A:
Matrix in scipy csr-sparse format of shape K x K where K = nVerts * outdim (Continuous Galerkin) or K = nElems * (dim+1) * outdim (Discontinuous Galerkin).
- nlfem.stiffnessMatrix()
Computes the stiffness matrix corresponding to the nonlocal operator
\[-\mathcal{L}(\mathbf{u})(\mathbf{x}) = 2 \int_{B_{\delta}(\mathbf{x}) \cap \widehat{\Omega}}(\mathbf{C}_\delta(\mathbf{x}, \mathbf{y}) \mathbf{u}(\mathbf{x}) - \mathbf{C}_\delta(\mathbf{y}, \mathbf{x})\mathbf{u}(\mathbf{y})) d\mathbf{y},\]on a given mesh. The input parameters are expected to be dictionaries. Find more information on the expected content in the examples/Test2D/conf/testConfFull.py. This function is a wrapper for the function par_assemble.
- Parameters:
mesh – Dictionary containing the mesh information (i.e. the keys
"elements","elementLabels","vertices", and"vertexLabels"). The values in the arrays"elements","elementLabels"and"vertexLabels"are expected to be of datatypenumpy.int. The values in"vertices"are supposed to be of typenumpy.float64. Elements in the domain have positive labels. Elements in the nonlocal Dirichlet boundary have negative labels. For this purpose it does not matter which positive or negative number is used, and the kernels can depend on the element labels. The labels of the elements should be consistent with the vertex labels. In the case of Discontinuous Galerkin this means that the signs of the element labels and corresponding vertex labels coincide. In case of Continuous Galerkin this means that elements with negative label have only vertices with negative label.kernel – The kernel is assumed to be a dictionary containing the keys
"function", and"outputdim". The value of"function"is a string which contains the name of the kernel (e.g. “constant”). You find all available options in the function src/Cassemble.cpp/lookup_configuration(). The key"outputdim"describes the output dimension of the kernel function. In case of a scalar valued kernel this would be 1. In case of the kernel “linearPrototypeMicroelasticField” this would be 2 (find an example in examples/Test2D/testConfFull.py). Note, that the value of"outputdim"does not define the output dimension of the kernel, but describes it. The value has to be consistent with the definition of the corresponding kernel in src/model.cpp.configuration – Dictionary containing the configuration (find an example in examples/Test2D/conf/testConfFull.py).
- Returns:
Matrix A in scipy csr-sparse format of shape K x K where K = nVerts * outdim (Continuous Galerkin) or K = nElems * (dim+1) * outdim (Discontinuous Galerkin).
- nlfem.loadVector()
Computes load vector. The input parameters are expected to be dictionaries. Find more information on the expected content in the example/Test2D/testConfFull.py
- Parameters:
mesh – Dictionary containing the mesh information (i.e. the keys
"elements","elementLabels","vertices","vertexLabels"and"outdim"), where"outdim"is the output-dimension of the kernel. In this function the output dimension is read from"outdim", as no kernel is expected. The load cannot yet depend on label information.load – Dictionary containing the load information (find an example in testConfFull.py).
configuration – Dictionary containing the configuration (find an example in examples/Test2D/conf/testConfFull.py).
- Returns:
Vector f of shape K = nVerts * outdim (Continuous Galerkin) or K = nElems * (dim+1) * outdim (Discontinuous Galerkin)